Skip to main contentIgDesign is an in-vitro validated tool to redesign antibody CDRs in complex with antigens for improved binding affinity. It’s developed by Absci and trained on the Structural Antibody Database (SAbDab) retrieved on December 6, 2022.
If you need to optimize an antibody/VHH binder bound to an antigen, IgDesign is a great choice! We recommend running IgDesign with 1M sequences (1000 batches) and taking the top 100 designs based on the lowest ce_loss. The authors found somewhat greater success designing HCDR3 only compared to HCDR123.
Experimental Validation
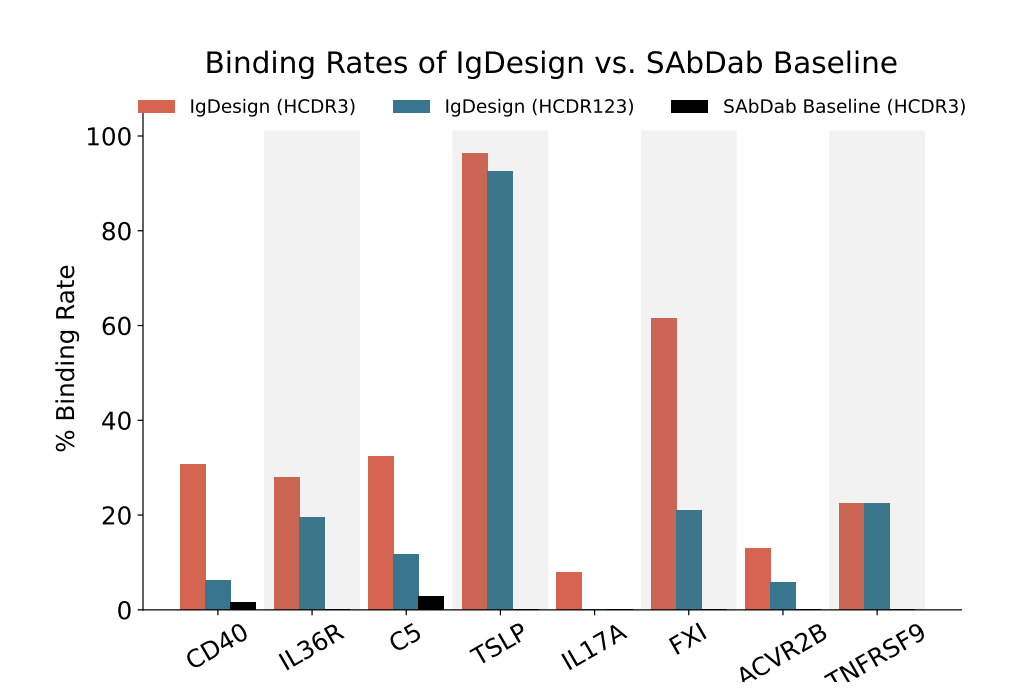
The following are binding rates assessed by SPR across antigens for IgDesign on HCDR3 and HCDR123, compared to SAbDab baseline with the native HCDR1 and HCDR2, and sampled HCDR3. 1437 antibodies were screened across 8 targets, and 278 binders were identified.
| Antigen | IgDesign (HCDR3) | IgDesign (HCDR123) | SAbDab (HCDR3) |
|---|
| Antigen 1 | 30.6% (22/72) | 6.3% (4/64) | 1.6% (1/61) |
| IL36R | 27.9% (19/68) | 15.6% (5/32) | 1.6% (1/68) |
| C5 | 53.8% (14/26) | 61.5% (8/13) | 16.7% (1/6) |
| TSLP | 96.3% (52/54) | 92.2% (47/51) | 88.1% (37/42) |
| IL17A | 56.3% (9/16) | 0.0% (0/8) | 12.5% (1/8) |
| FXI | 61.5% (32/52) | 49.0% (24/49) | 20.0% (1/5) |
| ACVR2B | 53.3% (16/30) | 9.4% (3/32) | 0.0% (0/9) |
| TNFRSF9 | 22.4% (15/67) | 24.3% (9/37) | 6.3% (2/32) |
- PDB File: Antibody-antigen complex structure in PDB format
- Heavy Chain ID
- Light Chain ID
- Antigen Chain ID
- Number of Batches: Number of batches to process, where each batches will generate 1000 sequences (recommended: 1000 batches)
- Verify Sequences: This will automatically submit your output sequences to a structure predicton tool for verification
- Select Custom CDR Indices: Optionally, select indices to design for each CDR instead of detecting them (IMGT numbered CDRs used by default)
Outputs
- Redesigned CDRs: A list of redesigned CDRs
- CE Loss Scores: Cross-entropy loss scores for each design, used to rank the designs (lower is better)
If you don’t have a antibody/antigen complex, you can try first predicting a docked structure of your complex using one of our structure prediction tools. If you have epitope information, you can incorporate restraints into your prediciton.
Try IgDesign 