Skip to main contentGuide: Working with Tamarind Bio Databases
Tamarind Bio’s software suite provides scientists and researchers with powerful tools to accelerate research and discovery. The database functionality offers a solution for organizing, categorizing, annotating, and comparing massive protein lists, enabling fast searching and insights on target sequences.

Database Walkthrough: Importing an Antibody List
The following steps outline how to import a list of antibodies into your Tamarind Bio database using a CSV file.
1. Accessing the Database
- Generate your CSV document containing your protein/antibody data.
- Click Database from the left-hand panel. This page displays all of your organization’s protein/antibody databases, including groupings, and allows for searching and filtering.
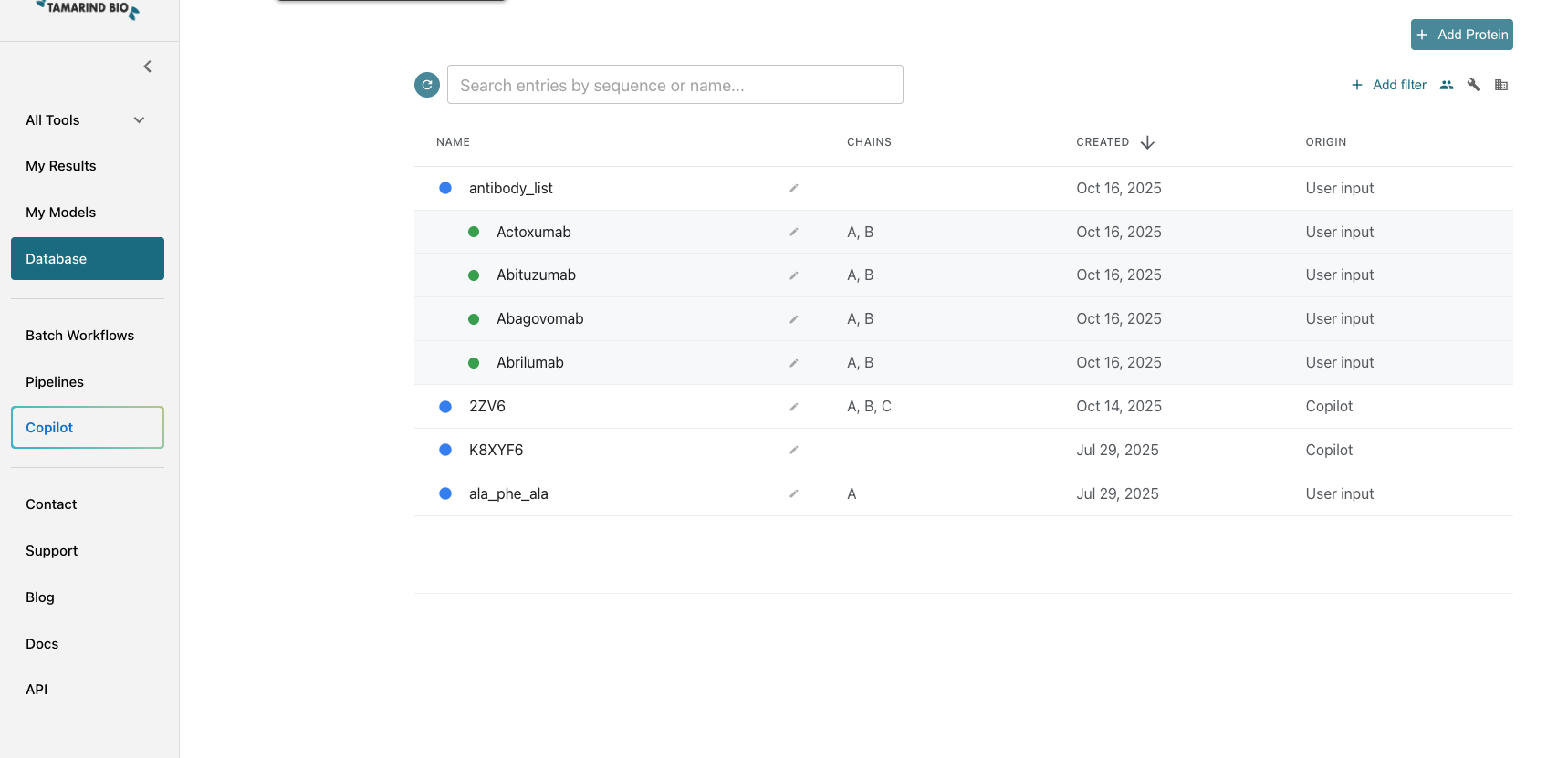
2. Starting the Import Process
- Click the green Add Protein button in the top right.
- Select Add Antibodies. (This option pre-checks specific fields for antibody data.)
- Click Add Protein List.
- Click the CSV tab to upload your data.
3. Uploading Your Data
- Drag & Drop or Browse for your CSV file, or select a file already uploaded into the Tamarind platform.
- Upload your file.
- Optional: Choose whether to deduplicate sequences and save count.
4. Defining the Database Group
- Name the Group: Choose a name for your list (e.g., Antibody Therapeutic).
- Select Sequence Column(s): Select the column(s) in your CSV that contain the sequence data (e.g., Heavy Sequence & Light Sequence).
- Group Name: Provide a unique group name (e.g., Sample_Antibody_list).
- Select the Sequence: Choose from the populated sequences. You can add more by clicking the ”+” sign.
5. Defining Parameters (Chains & Residues)
- Define Chains: Set parameters (e.g., select Heavy for Chain A and Light for Chain B).
- Options: You can rename chains, select specific residues, modify the sequence, and indicate if the sequence is a target or a binder.
- Add additional chains by clicking “Add Chain”.
- Sequence Verification: The next section displays all the Amino Acid sequences and other imported data for review.
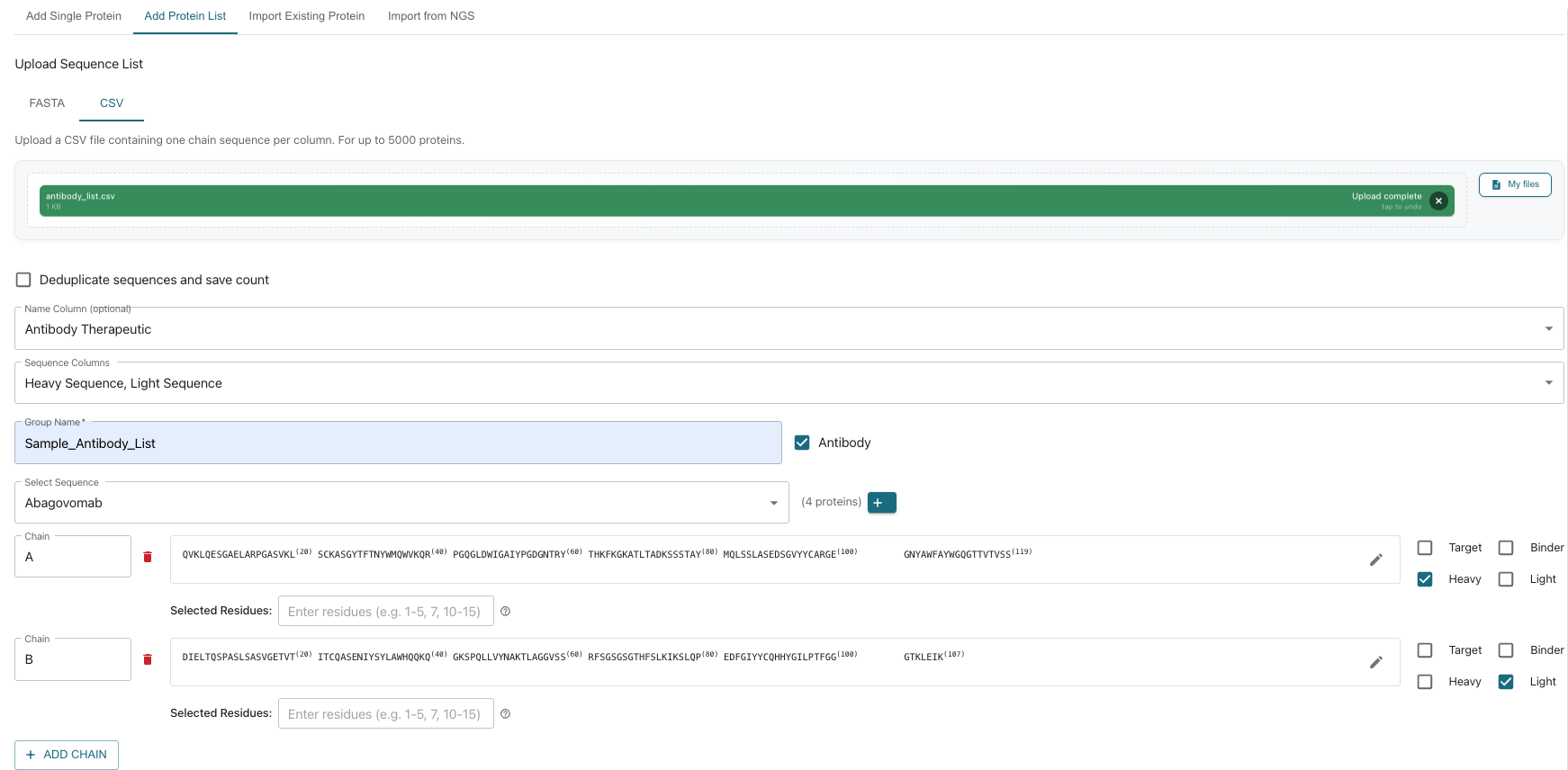
6. Adding Notes and Selecting Job Type
- Add Notes: Include any relevant notes on this protein group (e.g., specifics on generation, developer).
- Select Job Type (Analysis Models): Choose the analysis models. For antibodies, immuneBuilder, antibody-annotation, and HMMalign are pre-selected.
7. Finalizing and Reviewing
- Click Add Protein in the bottom right.
- Return to the main database page and click on the group name, “Sample_Antibody_List”, to view and analyze your data.
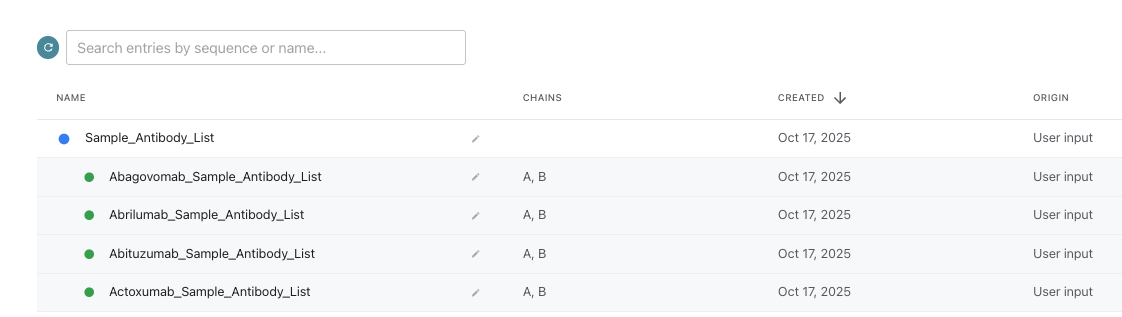
Analyzing Your Antibody Database Group
Once your group is imported, clicking on it opens a detailed analysis page. On the right side of the screen, you can quickly navigate to the various analysis sections via the Protein Table Navigation by clicking the teal <:
- Group Management: Add additional proteins, duplicate, delete, edit, or import more proteins into the list.
- Analysis Tools:
- Select PTMs (post-translational modifications).
- Identify clusters with threshold modifications.
- Score proteins using numerous scoring tools.
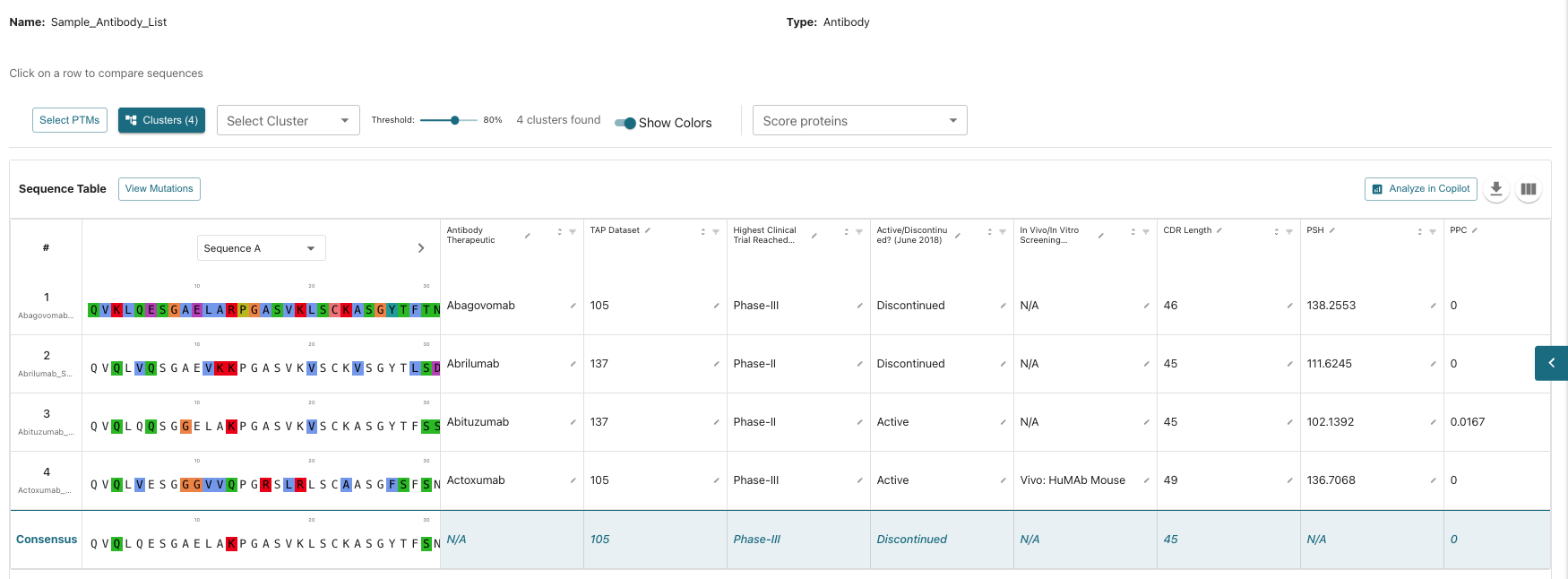
Sequence and Annotation Views
- Sequence Table: View amino acid sequences, identify mutations, compare point mutations, and scroll through sequences with coloring.
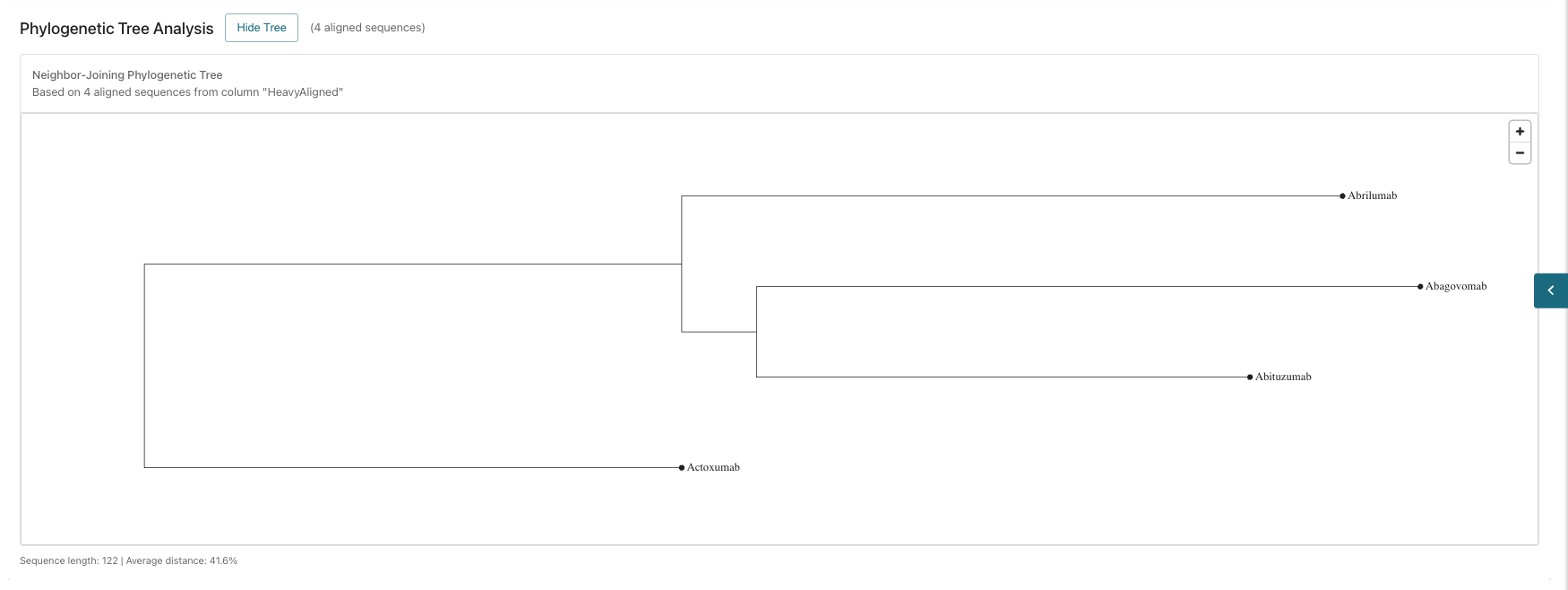
- Phylogenetic Tree Analysis: View the analysis for sequence comparison.
-
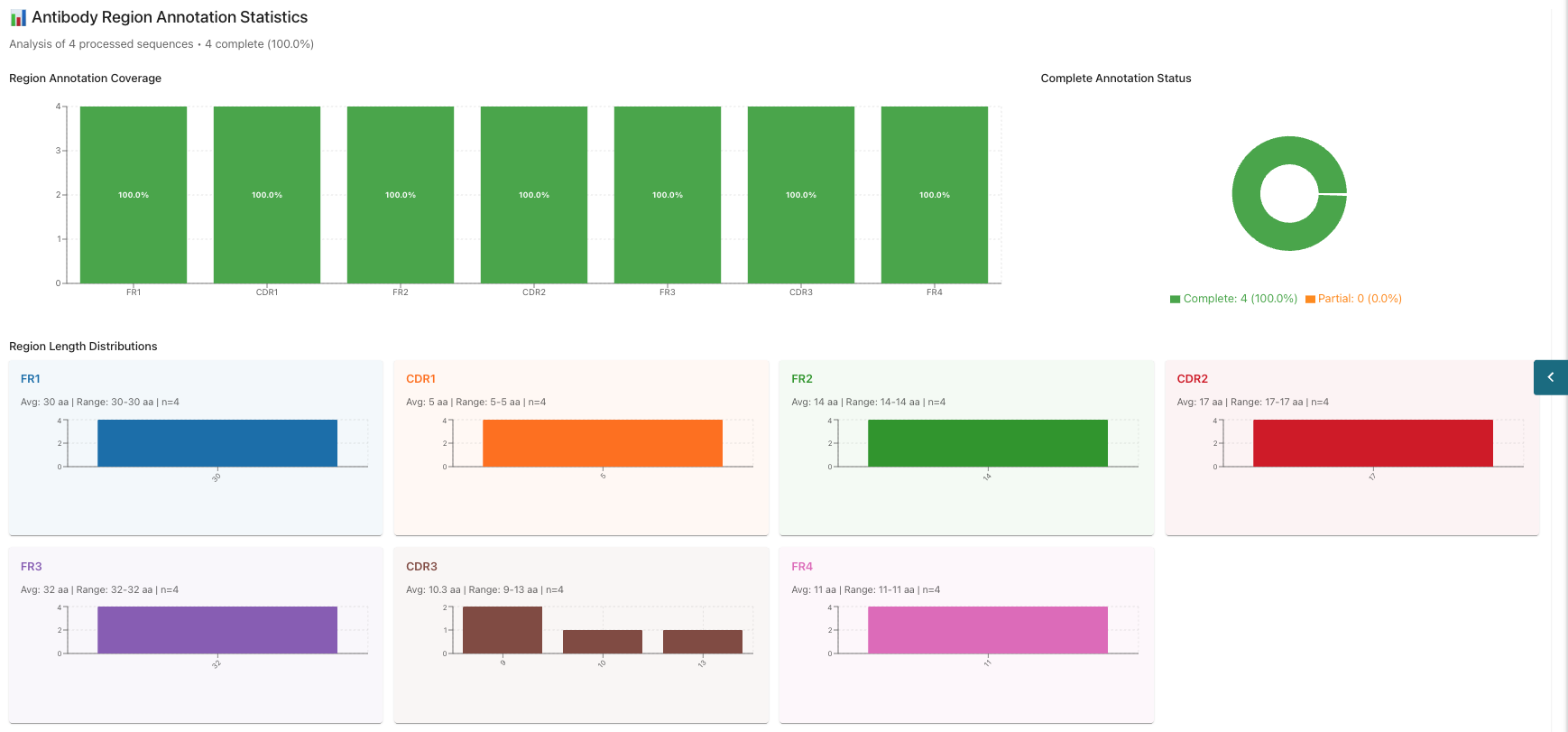
Annotation: View sequence annotations (check job status below the visualization, as generation may take a few minutes).
- In the annotation section, you can view statistics such as Region Annotation Cover, Complete Annotation Status, and Region Length Distribution.
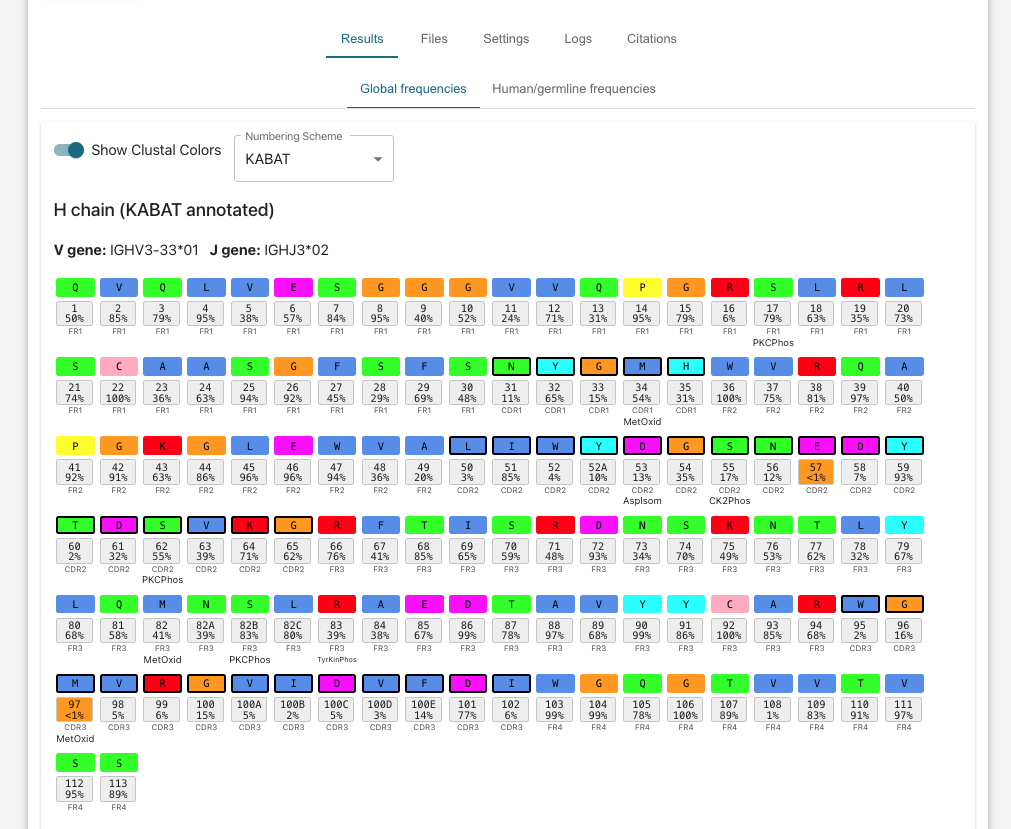
- Click into individual jobs in the Antibody Annotation window at the bottom to view color coded codes annotation of each individual amino acid with position occurrence frequency statistics.
-
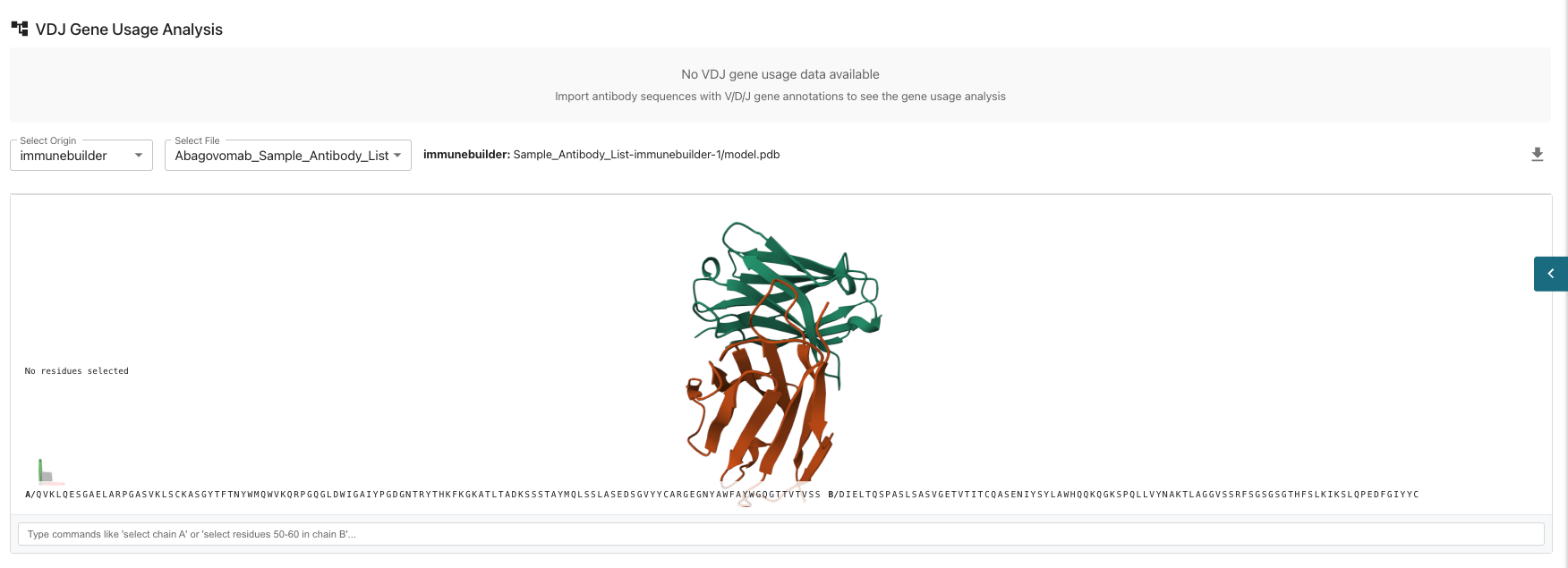
VDJ Gene Usage Analysis This involves studying the frequencies of Variable (V), DIversity (D), Joining (J) gene segments in the immune system, particularly in B-cell receptors, to understand antibody diversity and immune response. If you desire to use this analysis feature, make sure to import antibody sequences with V/D/J gene annotations to see the gene usage analysis.
-
Individual Sequence Visualizations: View your predicted structure using Immunebuilder where you can select individual amino acids or chains for further analysis. You can zoom into the individual amino acid chains to visually select specific chains of interest.
 The flexibility in inputs, models, and parameters allows for extensive research possibilities. It is recommended to test multiple sets of inputs, models and sequence inputs to compare results.
The flexibility in inputs, models, and parameters allows for extensive research possibilities. It is recommended to test multiple sets of inputs, models and sequence inputs to compare results.
Need Assistance?
If you have any questions about the database process or how to maximize the platform’s potential, feel free to reach out to the Tamarind Bio team:








 The flexibility in inputs, models, and parameters allows for extensive research possibilities. It is recommended to test multiple sets of inputs, models and sequence inputs to compare results.
The flexibility in inputs, models, and parameters allows for extensive research possibilities. It is recommended to test multiple sets of inputs, models and sequence inputs to compare results.